On the use of experimental observations to bias simulated ensembles
/Jed W. Pitera and John D. Chodera.
J. Chem. Theor. Comput. 8:3445, 2012. [DOI] [PDF]
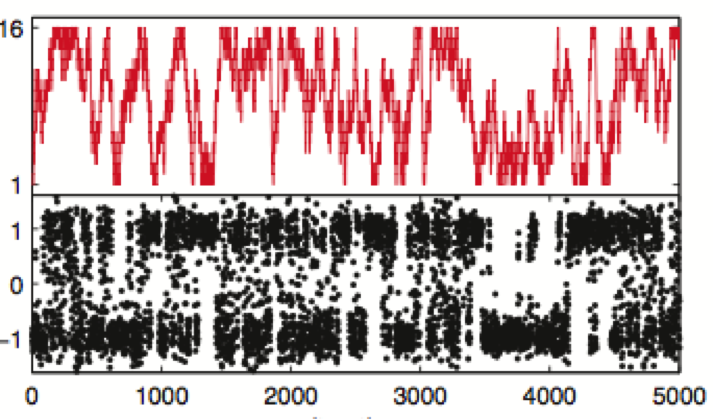
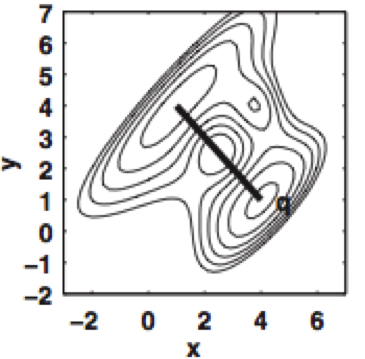
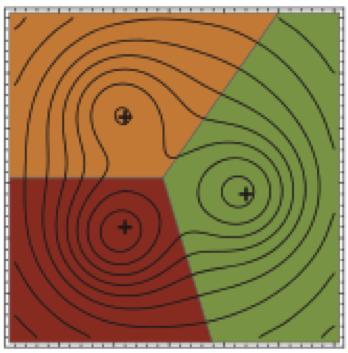
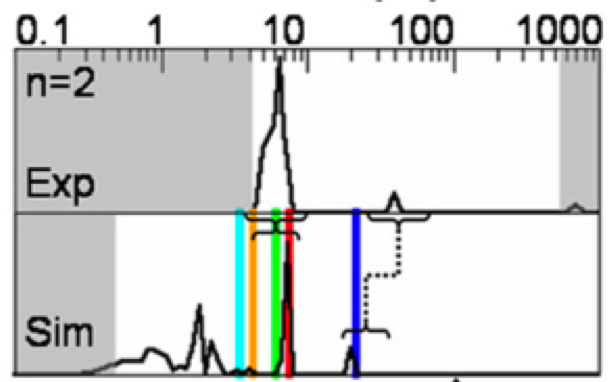
We show how the concept of maximum entropy can be used to recover unbiased conformational distributions from experimental data, and how this concept relates to the popular `ensemble refinement' schemes for NMR data analysis.